Cytoscape was originally created at the Institute of Systems Biology in Seattle in 2002. Now, it is developed by an international consortium of open source developers. Cytoscape was initially made public in July, 2002 (v0.8); the second release (v0.9) was in November, 2002, and v1.0 was released in March 2003.
| Original author(s) | Institute for Systems Biology |
|---|---|
| Developer(s) | Cytoscape Team |
| Initial release | July 2002 |
| Stable release | 3.8.2 / 29 October 2020; 5 months ago[1] |
| Written in | Java |
| Operating system | Any (Java-based) |
| Type | Image processing |
| License | LGPL |
| Website | www.cytoscape.org |
Cytoscape is an open sourcebioinformaticssoftware platform for visualizingmolecular interaction networks and integrating with gene expression profiles and other state data. Additional features are available as plugins. Plugins are available for network and molecular profiling analyses, new layouts, additional file format support and connection with databases and searching in large networks. Plugins may be developed using the Cytoscape open Java software architecture by anyone and plugin community development is encouraged.[2][3] Cytoscape also has a JavaScript-centric sister project named Cytoscape.js that can be used to analyse and visualise graphs in JavaScript environments, like a browser.
Cytoscape Manual
History[edit]
- Cytoscape is an advanced application designed for biology students, researchers and enthusiasts that want a reliable way to view and analyze molecular interaction networks and biological pathways. The tool also provides numerous ways to annotate your files, and wraps all of its features in a neatly organized interface.
- Cytoscape is an open source software project for integrating biomolecular interaction networks with high-throughput expression data and other molecular states into a unified conceptual framework. Although applicable to any system of molecular components and interactions, Cytoscape is most powerful w.
- Tutorial:Introduction to Cytoscape 2. At lower right is the Data Panel which can be used to display node, edge, and network attribute data The Network Management and Data browser panels are dockable tabbed panels known as CytoPanels.
Cytoscape was originally created at the Institute of Systems Biology in Seattle in 2002. Now, it is developed by an international consortium of open source developers. Cytoscape was initially made public in July, 2002 (v0.8); the second release (v0.9) was in November, 2002, and v1.0 was released in March 2003. Version 1.1.1 is the last stable release for the 1.0 series. Version 2.0 was initially released in 2004; Cytoscape 2.83, the final 2.xx version, was released in May 2012. Version 3.0 was released Feb 1, 2013, and the latest version, 3.4.0, was released in May 2016.
Development[edit]

The Cytoscape core developer team continues to work on this project and released Cytoscape 3.0 in 2013. This represented a major change in the Cytoscape architecture; it is a more modularized, expandable and maintainable version of the software.[4]
Usage[edit]
While Cytoscape is most commonly used for biological research applications, it is agnostic in terms of usage. Cytoscape can be used to visualize and analyze network graphs of any kind involving nodes and edges (e.g., social networks). A key aspect of the software architecture of Cytoscape is the use of plugins for specialized features. Plugins are developed by core developers and the greater user community.
Features[edit]
Input
- Input and construct molecular interaction networks from raw interaction files (SIF format) containing lists of protein-protein and/or protein–DNA interaction pairs. For yeast and other model organisms, large sources of pairwise interactions are available through the BIND and TRANSFAC databases. User-defined interaction types are also supported.
- Load and save previously-constructed interaction networks in GML format (Graph Modelling Language).
- Load and save networks and node/edge attributes in an XML document format called XGMML (eXtensible Graph Markup and Modeling Language).
- Input mRNA expression profiles from tab- or space-delimited text files.
- Load and save arbitrary attributes on nodes and edges. For example, input a set of custom annotation terms for your proteins, create a set of confidence values for your protein–protein interactions.
- Import gene functional annotations from the Gene Ontology (GO) and KEGG databases.
- Directly import GO terms and annotations from OBO and Gene Association files.
- Load and save state of the cytoscape session in a cytoscape session (.cys) file. Cytoscape session file includes networks, attributes (for node/edge/network), desktop states (selected/hidden nodes and edges, window sizes), properties, and visual styles.
Visualization
- Customize network data display using powerful visual styles.
- View a superposition of gene expression ratios and p-values on the network. Expression data can be mapped to node color, label, border thickness, or border color, etc. according to user-configurable colors and visualization schemes.
- Layout networks in two dimensions. A variety of layout algorithms are available, including cyclic and spring-embedded layouts.
- Zoom in/out and pan for browsing the network.
- Use the network manager to easily organize multiple networks. And this structure can be saved in a session file.
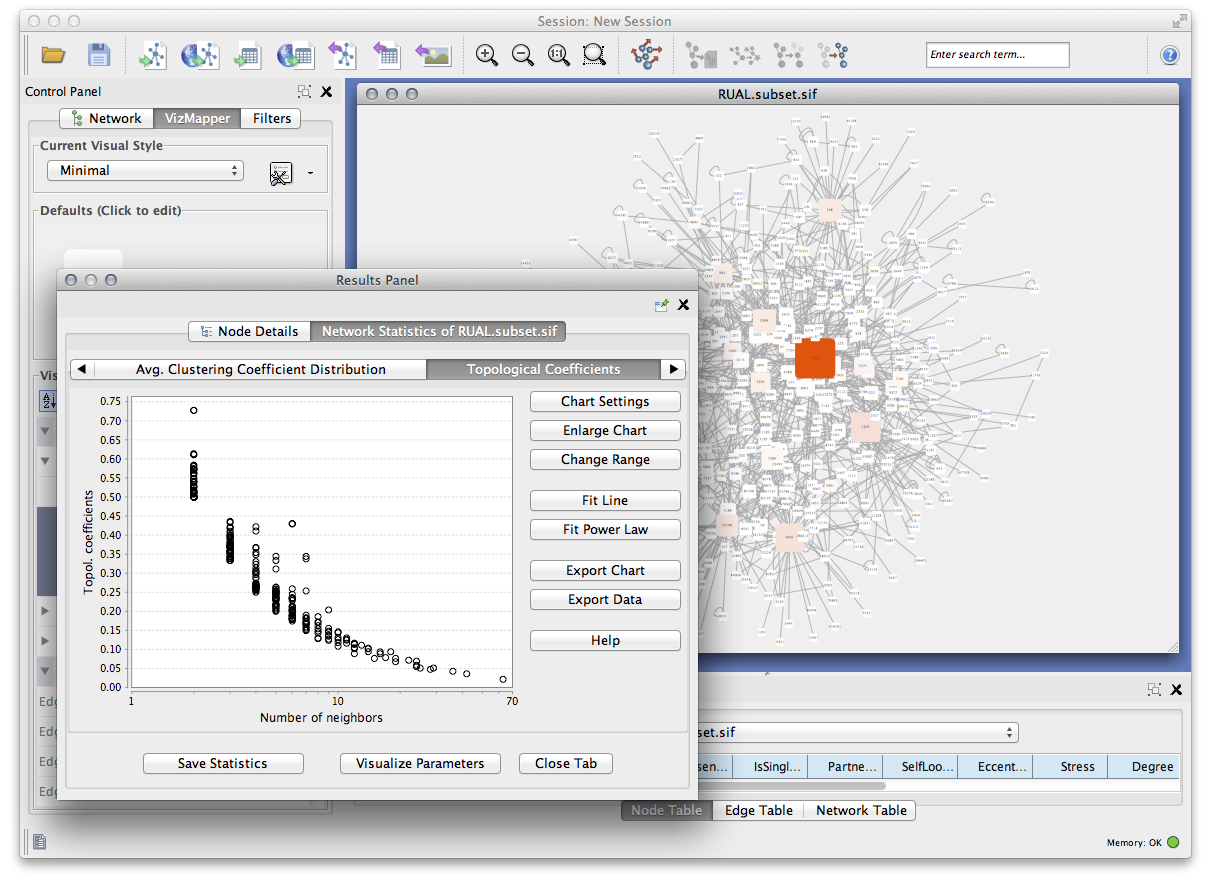
- Use the bird's eye view to easily navigate large networks.
- Easily navigate large networks (100,000+ nodes and edges) by an efficient rendering engine.
Analysis
- Plugins are available for network and molecular profile analysis. For example:
- Filter the network to select subsets of nodes and/or interactions based on the current data. For instance, users may select nodes involved in a threshold number of interactions, nodes that share a particular GO annotation, or nodes whose gene expression levels change significantly in one or more conditions according to p-values loaded with the gene expression data.
- Find active subnetworks/pathway modules. The network is screened against gene expression data to identify connected sets of interactions, i.e. interaction subnetworks, whose genes show particularly high levels of differential expression. The interactions contained in each subnetwork provide hypotheses for the regulatory and signaling interactions in control of the observed expression changes.
- Find clusters (highly interconnected regions) in any network loaded into Cytoscape. Depending on the type of network, clusters may mean different things. For instance, clusters in a protein–protein interaction network have been shown to be protein complexes and parts of pathways. Clusters in a protein similarity network represent protein families.
See also[edit]
References[edit]
- ^'Cytoscape 3.8.2 is released!'. groups.google.com. Retrieved 11 March 2021.
- ^Shannon P, Markiel A, Ozier O, et al. (2003). 'Cytoscape: a software environment for integrated models of biomolecular interaction networks'. Genome Res. 13 (11): 2498–504. doi:10.1101/gr.1239303. PMC403769. PMID14597658.
- ^Bell GW, Lewitter F (2006). 'Visualizing networks'. Meth. Enzymol. 411: 408–21. doi:10.1016/S0076-6879(06)11022-8. PMID16939803.
- ^Cytoscape Product Roadmap
External links[edit]
For this tutorial, please use this node list.
As we learned in the last tutorial, all Cytoscape really needs is an edge list with two columns in it. But you might also want to supply Cytoscape with a list of nodes. That way, you can feed Cytoscape extra information that you can use to distinguish among nodes. That extra information about your nodes is called attributes.
(Actually, edges can have attributes, too, as you might have guessed. In the case of edges, the attributes usually describe the nature of the connection between nodes. For example, if I had a People column and a Book column, I could use edge attributes to describe the nature of each connection: published, wrote, illustrated. We’ll just deal with node attributes here, but the same steps apply to edge attributes, too.)
Preparing a node list
Your node list should contain, at a minimum, one column that supplies the name of every node once. My edge list, as you may have noticed, contained many actors’ names and film titles multiple times. For example, since Ida Anderson appeared in multiple films, I had rows that looked like this:
| actor | film |
|---|---|
| Ida Anderson | Deceit |
| Ida Anderson | A Son of Satan |
| Ida Anderson | The Secret Sorrow |
| Ida Anderson | Gayety |
| Ida Anderson | Ghost of Tolson’s Manor |
We don’t want this in our node list, though. We only want Ida Anderson to appear once, and we only want each film’s title to appear once. Actors and films should all be in the same column.
By convention, we usually label node names id, although it won’t mess anything up if you want to label the column something else.
In subsequent columns, you can provide attributes for each of your nodes. I’ll just supply one here, called type, containing either actor or film. You could provide multiple columns, though, like gender or director.
If you have an edge list but no node list, here is how to convert an edge list into a node list.
For this tutorial, please use this node list.
Add your node list to your Cytoscape graph
Head back to Cytoscape, where the graph you created during the previous tutorial should be open. We’re now going to add our new node list to the existing graph.
Do that by clicking on the Import Table from File button, circled below. In the window that pops up, select your node list and click Open.
Check to make sure Cytoscape interpreted your data properly
Hopefully, the window that pops up looks something like the one below. The column labeled with a key is the column Cytoscape will use to try and match your node attributes with the nodes that already exist in your Cytoscape graph.
You can see that Cytoscape has labled your type column with an icon that looks like a document. That means that Cytoscape has interpreted that column, correctly, as attribute information for your nodes. This all looks good, so we can click OK.
What happened?
Not a lot, at first glance! But if you look at the pane at the bottom of Cytoscape’s window and click on the Node Table tab, you’ll see that your node table now has an extra column, in which each entity is labeled as either an actor or a film.
That’s good! It means the last step worked!
Use node attributes to color nodes (1)
Now that Cytoscape knows which nodes are which, we can color the nodes based on whether they’re an actor or a film. To get started, click on the Style tab in the control panel, and make sure the Node tab is selected at the bottom of that pane. Your control panel should look something like mine in the image below.
Use attributes to color nodes (2)
Remember, the Map. column is the one we’ll use to color nodes based on their attributes. Click on the Map. button in the Fill Color row. You should now see some additional options.
For Column, choose type. (If you included additional attributes in your node list, here’s where you could select those as the basis of your nodes’ colors.)
For Mapping Type, choose Discrete Mapping. That means that each node type will receive a different color of your choosing. (Passthrough mapping is for when you’ve supplied visual attributes, like colors, directly in your node list.) Some properties offer you a Continuous Mapping option. That’s for cases when nodes don’t differ in type, but in degree — for example, in cases where you’d like your nodes to increase in size based on how many connections they have. But we’re not using that kind of data right now.
Give each type of node a color by clicking on the tiny button that contains three dots. (It appears just the left of the garbage can.) Your graph should change color to reflect your choices.
Using Cytoscape’s network analysis tools to calculate attributes
You can supply your own attributes to Cytoscape, as we just did with our node list. However, part of the power of Cytoscape is its ability to calculate certain attributes of your network for you. For example, Cytoscape can provide each node with a number that reflects its degree, meaning the number of connections to and from that node. We can then use that number to control the size of our nodes.
To get Cytoscape to provide these values, choose Tools from Cytoscape’s menu, then Network Analyzer, then Network Analysis, and finally Analyze Network. In the window that pops up, Cytoscape will ask you whether you’d like it to treat your edges as directed or undirected. Remember, all of our connections are reciprocal, so select Treat Network as Undirected. Once you click OK, Cytoscape will perform some calculations and fill in some new columns in your Nodes, Edges, and Network tables.
Understanding these statistics
To understand what these numbers mean, refresh your memory of the social network analysis glossary I provided you in the last tutorial. Here’s an important caveat, though: Some of these statistics are meaningful for the kind of graph we have, and some of them aren’t. We have a bimodal network, meaning we have two different kinds of things: actors and nodes. Not all measures of networks make sense for bimodal networks.
Degree centrality, for example, still makes some sense for bimodal networks. If a node has a bunch of connections, we know it’s either an actor who appeared in a lot of movies or a film that contains a lot of actors. (Although, obviously, those two qualities are two very different things.) The clustering coefficient statistic, however, means very little for bimodal networks, because your network contains two different orders of things. To read more about measures of bimodal networks, see Scott Weingart’s stern words of advice.
Use your statistics to control the size of nodes
OK! We’ve calculated some statistics for our nodes, and we’ve been sternly lectured about the meaning (or lack thereof) of these measures. Let’s now (judiciously!) use our measures to control the visual appearance of our network graph.
Cytoscape Software
From your control panel, make sure the Style pane is selected, and then make sure the Nodes tab is selected at the bottom of the window. This time, click on the Map. button for the Size property. From the Column menu, select Degree (because, remember, degree still makes a certain amount of sense for bimodal graphs).
For Mapping Type, select Continuous. Remember, this means we’re distinguishing among our nodes along a spectrum, not breaking them into groups according to type. (We could use Discrete Mapping though! For example, all nodes that have five or fewer connections could be tiny and all nodes that have six or more connections could be big. It just depends on what you want to show.)
You can, if you wish, fiddle with the Continuous Mapping Editor by double-clicking the histogram that appears beneath the Continuous Mapping option. This allows you to control the maximum and minimum size of your nodes, along with the break points, if any.
Cytoscape Layout
Taking stock
We’ve done a lot! We’ve learned how to import a node list; how to use node attributes to control the size, shape, and color of our nodes; how to calculate network statistics; and how to use those statistics to control the appearance of our graph. We can now derive more meaning from the graph we’re working with; for example, it’s easy, at a glance, to get a sense of which actors appeared in the most films, and which films employed the most actors.
Our graph is still pretty overwhelming for the viewer, though. In the next tutorial, we’ll learn how to work with just parts of it at a time.
